UCloud is not just an ideal platform for the individual researcher who wants interactive access to HPC resources or an easy way to collaborate with national or international partners. It is also highly suitable for teaching. Jennifer Bartell and Samuele Soraggi, who are both working on the project National Health Data Science Sandbox for Training and Research, share their experiences with using UCloud.
National “sandbox” platform
The growing amounts of data in all research fields offer researchers new opportunities and possibilities for scientific breakthrough. In the case of health science, the use of large amounts of data has great potential to improve our health care – it can e.g. expand our ability to understand and diagnose diseases. One of the constraints of using health data is that many datasets (e.g. person-specific health records or genomics data) are sensitive from a patient privacy perspective and governed by strict access and usage guidelines. This can be a major challenge in particular for students or researchers who are just learning best practices in handling health data while also developing data science skills.
The idea behind the project National Health Data Science Sandbox for Training and Research is to provide students and researchers with a “sandbox” environment where they can develop their skills, test their ideas and learn how to analyze large biomedical and clinical datasets in an HPC setting. The national sandbox will contain only public, anonymous data and non-person-sensitive synthetic/simulated data which can be used without having to worry about GDPR regulations. It will also contain learning modules that guide trainees through major areas of health data science with a focus on new technologies and tools, simultaneously advancing their HPC skills as well as their grasp of the latest single-cell RNA-Seq analysis workflow, for example.
“We provide the tools and non-sensitive data sets for people to learn and test new tools and algorithms without having to worry about working with sensitive health data which adds tons of restrictions and constraints on what you do,” explains Jennifer Bartell, who is the coordinator of the project and a data scientist.
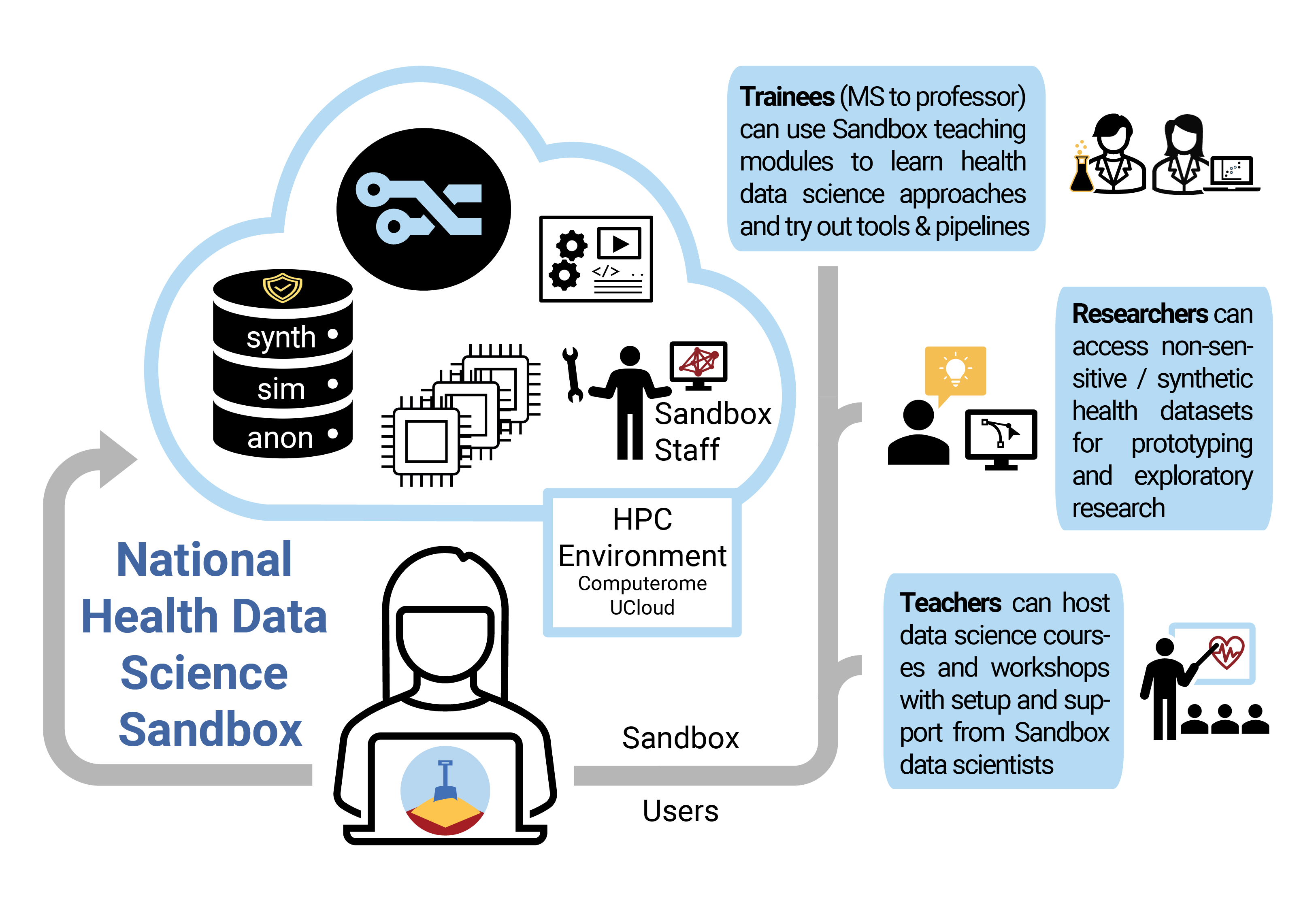
The Sandbox team is researching how to safely develop useful, privacy-preserving synthetic data as well as leverage published datasets. Their aim is that researchers and students with interesting or promising Sandbox test results can easily transition their approach and tools to sensitive datasets – with more evidence that their applications for data access and additional security measures are worth pursuing!
The National Health Data Science Sandbox is funded by the Novo Nordisk Foundation and lead by Professor Anders Krogh from the University of Copenhagen. The Sandbox is being built and supported by a consortium of data scientists from the universities of Copenhagen, Aarhus, Aalborg, Southern Denmark and the Technical University of Denmark.
UCloud as part of the Sandbox
The National Health Data Science Sandbox is using the secure cloud technologies available at the Computerome2 installation in Roskilde and the UCloud platform, originally developed by the University of Southern Denmark and now used as the basis for the DeiC Interactive HPC service. Using already existing facilities will ensure that students and researchers are accustomed to the systems that they may subsequently use for projects which involve real sensitive data.
This summer, UCloud was used for a course, Introduction to Next Generation Sequencing Data, which was organised in connection with the National Health Data Science Sandbox project.
“The Sandbox is a project supporting teaching and training in health data science, and we are running training ourselves in how to use the Sandbox / HPC environments to perform health data science. We are packaging Sandbox datasets and tools into topical training modules which have self-tutorial, stand-alone versions but which we also use to support in-person workshops. This is how we’re refining the material we provide in the Sandbox,” says Jennifer Bartell.
“UCloud is pretty ideal for organizing pedagogic material because it is interactive. The user support team at the eScience Center helped us so we basically had an app running and starting the packages so the users would not have to install anything and could directly use the course material interactively,” says Samuele Soraggi, also a data scientist working on the sandbox, and who, together with researchers at Aarhus University, organized the course.
Jennifer Bartell also expresses her satisfaction with UCloud and the support the project received from the eScience Center’s staff:
“It has been really nice working with Emiliano Molinaro and Claudio Pica. They have been really responsive and engaged in helping us roll out these education-focused apps on UCloud. We are happy to be working with partners that are excited about supporting teaching and training in an HPC environment,” says Jennifer Bartell.
The course has been published as an app on UCloud, Genomics Sandbox, which can be used by other researchers organizing similar courses or for independent study by any student or researcher. Course materials can thus be applied to new datasets uploaded by users. Each course has a companion webpage with additional material (slides, explorable code, and notes, etc) that is hosted on the Sandbox webpage and linked to in each app’s UCloud documentation.
“I think it’s very nice that the course participants did not have to worry about packages and installation. They could also upload their own datasets and try out the same analysis for their research projects. This allowed us to mostly concentrate on the tutorial for the course,” says Samuele Soraggi.
More recently, UCloud was also used for a bulk RNA-Seq workshop developed by the Copenhagen University Sandbox staff in collaboration with the HeaDS DataLab, which took place on the 18th and 19th of August. The in-person workshop had 26 participants and, as with the Introduction to Next Generation Sequencing Data course, the reviews were quite positive. This material will soon be made available as the Transcriptomics Sandbox app on UCloud. A Proteomics Sandbox app is also under active construction with associated workshops rolling out in 2023.
All course material and associated datasets are open source and available through GitHub and the Sandbox website, where future courses and training modules hosted at UCloud or Computerome will also be advertised.
Would you like to host a similar course on UCloud?
If you have course material and might want help implementing it in the Sandbox or if you would like the Sandbox team’s assistance in running a course or workshop related to health data science, you can contact Jennifer Bartell at NHDS_sandbox@sund.ku.dk or the Sandbox data scientist located at your university (listed here).
“If anybody has a course related to health data science, which they think would be fitting to host on UCloud in this way, but they maybe do not have the time or the resources to set it up, we would be happy to discuss deployment options in the Sandbox,” says Jennifer Bartell.
“It’s important to say that we also support the course when it’s running, both at a Slack-channel level, but we also have people on site at five universities who can help you,” says Samuele Soraggi.
UCloud can also be used for hosting workshops and courses in all research disciplines. If you need help to use UCloud for your course, please contact the support team: support@escience.sdu.dk.
Find out more
You can read more about the National Health Data Science Sandbox for Training and Research here and visit the project’s website here.
Prof. Anders Krogh, University of Copenhagen, who is the leader of the project, presents the concept of the National Health Data Science Sandbox in a video here.
UCloud has also been used in a similar way for a workshop organized by the Royal Danish Library. You can read both the participants’ and the organizer’s reflections here and here.